To some people, high-throughput screening (HTS) is synonymous with finding hits. But is it the best and only system? More and more drug researchers say they are looking for a faster and more cost-effective replacement. LeadBuilder was developed as an alternative to HTS that saves time and money because only compounds likely to become a drug are screened. HTS works the opposite way round – it ploughs through an enormous, random library of compounds and the hits are tested afterwards to find the most 'drug-like'.
HTS is like searching for a needle in a vast haystack using a bulldozer. It is expensive because – paradoxically – the chance of finding the needle increases if you have more hay in which to look. Hundreds of thousands, or even millions, of compounds must be collected before HTS can begin. You also need to buy the bulldozer. Equipment like assay platforms and automated machinery must be purchased. HTS has high ongoing costs too, such as reagents, supplying proteins or cells, and replenishing the compound collection over time.
Once the equipment is running, screening seems very fast at first glance. Some estimates claim 10,000–100,000 compounds a day can pass through an HTS system. But preparing for HTS is an often forgotten time-sink. It takes time to establish a robust assay suitable for HTS and to obtain enough high-quality protein or cells to screen the whole compound collection.
Analysing HTS results is also time-consuming. Bulldozing your way randomly through any old compounds means hits often are not very ‘drug-like’. Hundreds of hits must be exhaustively tested for safety, bioavailability, novelty and affinity for the target.
A decade ago, it was less important that HTS was expensive and hard to set up. Big pharmaceutical companies with large budgets and laboratories did most drug discovery. Now, spin-off companies, small biotechs, research institutions and academic labs are increasingly getting in on the act. Many cannot afford to run an HTS operation in-house or pay perhaps £100,000–£250,000 to outsource a screen.
LeadBuilder turns HTS on its head. Instead of screening everything, the system physically screens only compounds that look like drugs or hits. Returning to our haystack idea, the system only picks short “needle-like” straws, making it easier to find the ones that really shine (see Fig. 1 for differences between the two hit finding processes).
Based on state-of-the-art virtual modelling and screening software, LeadBuilder has four main modules: LibraryBuilder, CompoundProfiler, ScreenBuilder and StructureBuilder (Fig. 2).
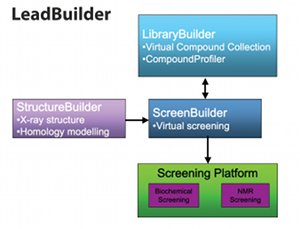
Fig. 2: The four main modules that make up LeadBuilder
The system starts with CATS (Compounds Available To Screen). These are a set of commercially available compounds sourced from reliable suppliers stored in the LibraryBuilder module.
ranked by drug potential
CompoundProfiler finds NICE (Interesting Chemical Entities) CATS by calculating each CATS’ molecular properties and predicting its physiochemical, pharmacokinetic and toxicity profile (Fig. 3).
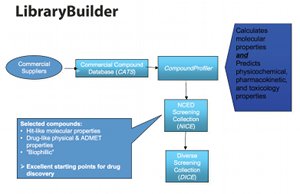
Fig. 3: Compounds are then ranked by how drug-like they are
CATS are then ranked by how ‘drug-like’ they are. NICE CATS meet every criterion for the ‘perfect’ screening hit – they have excellent molecular properties, good ADME, low toxicity and interact well with a target protein.
Choosing NICE CATS involves creative thinking about what makes a ‘drug-like’ compound. For example, after reviewing 37 hit-to-lead programmes, we found that high ligand efficiency (binding energy per atom) predicted hits that were easy to optimise into drug candidates. Higher ligand efficiencies resulted from certain structural features like charged groups (e.g. N+). So Domainex developed an algorithm that predicts a ligand efficiency from a compound’s structural features. We use this to eliminate poorly performing CATS.
The next stage in building leads is ScreenBuilder – a virtual screening engine that works on NICE CATS. Using information about the target protein, any known ligands or both of these, this module can screen by a combination of docking, pharmacophore searches and privileged fragment recognition. If the target protein is unknown, the system can identify a surrogate with a similar active site (Fig. 4).
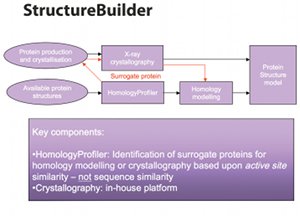
Fig. 4: Compounds with similar active sites can be identified
Three real-life examples can illustrate how ScreenBuilder works: first, screening with pharmacophores derived from a known ligand, followed by docking into the x-ray structure of a protein; second, searching with ligand-based pharmacophores; and third, using only information about the target protein structure.
Example 1: Finding small molecules to inhibit protein-protein or peptide-peptide interactions is notoriously difficult. But LeadBuilder managed it. ScreenBuilder found 47 compounds that blocked a growth factor – natural peptide – from binding to its protein receptor. Aberrant growth factor signalling often drives tumour growth.
Domainex was given an x-ray structure of the ligand-binding pocket of the growth factor receptor with an oligopeptide fragment of the growth factor ligand bound to it. LeadBuilder analysed the ligand-binding pocket to define search criteria. Several structural features of the oligopeptide – H-bond donors and acceptors and a lipophilic group – seemed essential for binding to the receptor.
Next, the NICE database was virtually screened for compounds that contained these features and could be ‘docked’ into the protein’s active site. Domainex’s client lined up a subset of these compounds for further screening.
The project is currently in its hit-to-lead phase. Altogether, LeadBuilder identified 950 compounds for screening, delivering a 5% hit rate over several structural types. This made it 10 times more productive than another vendor’s focused library of 1000 compounds, which found only five hits (0.5% hit rate).
Example 2: A patent was filed on several compounds identified during another LeadBuilder project because they had commercially interesting activity levels. Domainex had been just given one known ligand to work with. The client was interested in compounds that would stimulate bone growth to accelerate the healing of fractures. It had developed a cell-based assay to look for compounds that switched on a signalling pathway that would cause deposition of bone. The ligand given to Domainex as a starting point was the only stimulator it had found for the pathway – it was keen to find others as potential drugs and for mechanism-of-action studies.
Domainex screened the NICE database using three pharmacophoric features based on the known ligand. Around 100 compounds were selected for screening, with the emphasis being on cellular penetration. During screening, our client found a hit with five times the activity of the known ligand. We are now helping it with a follow-up medicinal chemistry programme to find even better compounds.
Example 3: Protein kinases are important drug targets for a number of diseases. Many of the known inhibitors work by competing with the kinase substrate ATP. The problem with this approach is that it is difficult to find compounds that stop ATP binding to one kinase, but have no effect on the binding of ATP to other kinases. This selectivity problem means that kinase inhibitors often have side-effects caused by them blocking non-target kinase enzymes.
Our client wanted to find a selective inhibitor of a kinase that is a well-known cancer target, and had a novel hypothesis – did this protein kinase have a natural regulatory site that they could exploit with small-molecule drugs? The answer was yes. The client has patented the IP and is using it to secure funds. Domainex is now helping it develop a programme to turn its hits into leads.
An x-ray structure of the kinase showed that, in the naturally occurring inactive form, part of the enzyme ‘loops’ around to block the substrate protein binding site. On activation, this loop must move away to reveal a groove on the enzyme where the protein substrate can bind. An inhibitor that sits in this groove would stop catalysis by the kinase.
LeadBuilder found a four-point pharmaco-phore based on the key features of the “autoinhibitory loop” and an analysis of its binding groove on the surface of the enzyme (see Fig. 5). Using ScreenBuilder, potential ligands were selected from the NICE database using this pharmacophore and were docking into the hypothesised binding pocket.
Domainex recommended that the client screen 450 compounds based on this research. When tested against the kinase, this gave a 6% hit rate – 27 hits in four structural classes. Four of these showed promise in a follow-up cell-based screen.
In conclusion, dissatisfaction with HTS is growing. To many, finding hits seems like looking for a needle in a haystack. LeadBuilder is a smarter alternative. It may not yet offer pinpoint accuracy, but it can deliver ten times more hits during drug development.
