The Black Death, a bubonic plague that wiped out nearly two thirds of Europe's population in 4 years, still has up to 3000 annual cases worldwide.
Luckily, antibiotics can successfully treat most cases in 2–5 days, just as antibiotics can help to stave off infections for everything from routine wisdom tooth extractions to organ transplants and chemotherapy. However, this could all change, writes Chris Lock, Vice President of R&D at SCIEX.
The European Commission is reviewing its pharmaceutical policy framework this year and world leaders announced their commitment to tackle AMR at the last G7 Summit.1,2
Short of unrealistic expectations for the rapid development of new antibiotics, the scientific community must focus on combatting AMR, a mission that requires better, faster analysis of infection for more accurate antibiotic treatment to avoid further disease and increased resistance.
Doing so will also help to restore Europe’s position as an innovation leader in the discovery, development and delivery of new diagnostics, therapies and vaccines.
Analytical tools to support precision medicine with antibiotics
In addition to developing new approaches to accelerate the discovery and development of antimicrobials, researchers are also working on ways to better optimise the application of existing therapeutics to combat microbial disease.
Jerome Lemoine, PhD, a professor at the Institute of Analytical Sciences in Lyon, France, is investigating how diagnostic tools can be improved to help clinicians to make better informed decisions when treating life-threatening infections such as those caused by methicillin-resistant Staphylococcus aureus (MRSA), which can result in sepsis and ventilator-associated pneumonia (VAP).3,4
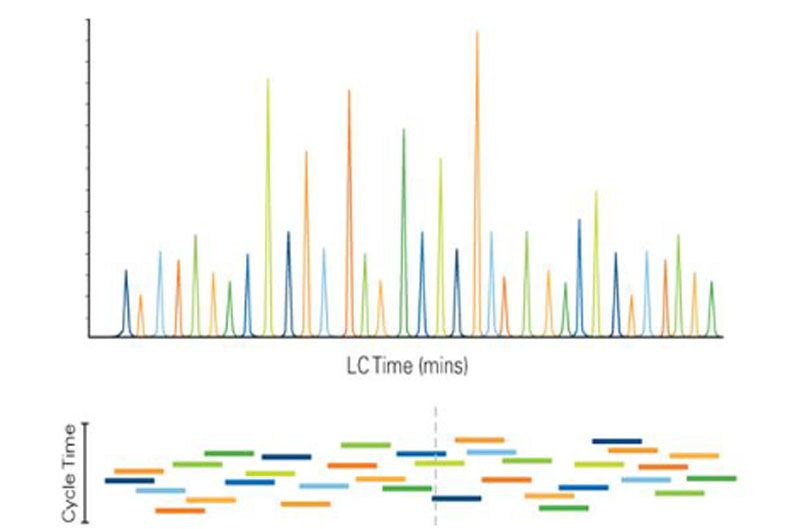
Figure 1: Scheduled MRM algorithm. Using knowledge of the elution time of each analyte, all MRM transitions are monitored only during short retention time windows. This allows many more MRM transitions to be monitored in a single LC run, while still maintaining maximised dwell times and optimised cycle times9
In collaboration with Prof. François Vandenesch of the Hospices Civils de Lyon, Hôpital de la Croix Rousse in Lyon, France, Prof. Lemoine is developing a new analytical method to inform and accelerate the identification of bacteria from clinical samples, including the characterisation of virulence and any antimicrobial resistance.3,4
MRSA is typically a hospital-acquired or “nosocomial” infection. It caused more than 100,000 deaths worldwide in 2019 and it’s only one of the many pathogenic bacteria that exist with AMR.5
Globally, about 1.27 million people died from bacterial AMR in 2019, with an additional 3.68 million deaths associated with bacterial AMR.5 Nosocomial infections such as sepsis and VAP are caused by a number of these pathogenic bacteria.
Identifying the bacterial species that has caused an illness can take time … and determining whether that particular strain is susceptible or resistant to specific antimicrobials can take even longer.3,4
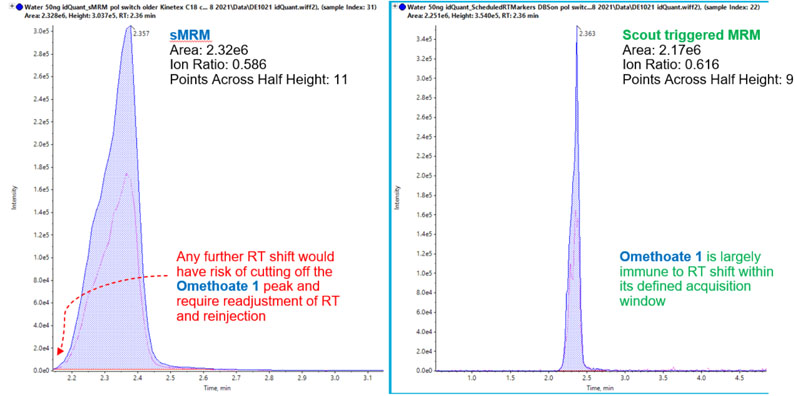
Figure 2: Omethoate using scheduled MRM versus Scout-triggered MRM. Data quality is comparable between the time-scheduled MRM and Scout triggered MRM for parameters such as peak area, ion ratio and points across half height. A slight retention time shift in the time-scheduled MRM trace risks this peak becoming cut off or missed (left), rendering the sample unusable and requiring reinjection. In contrast, the Scout triggered MRM peak safely measured as retention time shifts are automatically adjusted for8
Patients are usually treated empirically with a broad-spectrum antibiotic because treatment needs to begin as soon as possible; however, this practice can increase the risk of selecting for multidrug-resistant bacteria.4
Therefore, it is critical to identify the bacteria responsible in each patient’s case, confirm the diagnosis and treat with the antimicrobial known to be most effective against that bacterial strain.3,4
Developing improved mass spectrometry assays to identify pathogenic bacteria
Approximately 50% of patients taking antibiotics start by being prescribed the wrong one and without proper identification of the pathogenic bacterium.6 Fast, robust and affordable technologies and workflows are still needed for bacterial identification and characterisation, including antimicrobial susceptibility testing (AST).3,4,6

To help meet this need, Prof. Lemoine has spent years developing mass spectrometric methods that can provide fast and definitive analyses of bacteria from a variety of clinical samples (kindly donated from hospitals).3,4,7 Mass spectrometry (MS) has emerged as the gold standard technology for bacterial characterisation in clinical microbiology.3,4
In conjunction with our scientists at SCIEX, Prof. Lemoine has been developing a rapid workflow using MS and liquid chromatography (LC) with a view to one day implementing it in the clinical setting. This method uses software algorithms to acquire analytical data based on the detection of analyte triggers known as Scout marker compounds.7
This algorithm is being incorporated into the existing MS technique: multiple reaction monitoring (MRM). MRM is a highly specific and sensitive MS operation mode for targeted quantitative analysis that uses tandem MS (MS/MS) with LC.8
Prof. Lemoine is using targeted quantitative LC-MS/MS analysis because it is a multiplex approach that’s sufficiently selective, sensitive and robust enough to identify and characterise bacteria from biological samples — providing a wealth of information in a single run.
The evolution of MRM in research
To quantify an increasing number of target analytes per sample and per run, scientists incorporate retention time into the MRM method, so a full panel of analytes can be captured without compromising data quality.8 Using the scheduled MRM, analytes are assigned retention times during which the mass spectrometer scans for the analytes expected to elute from the LC column.
By adding a schedule of retention times, the sensitivity and selectivity are improved and the number of analytes that can be targeted in a single acquisition increases.8
This widely adopted, time-scheduled MRM strategy allows for the analysis of hundreds and even thousands more transitions in a single method, while maintaining reasonable cycle and dwell times for quantitative data quality (Figure 1).8,9
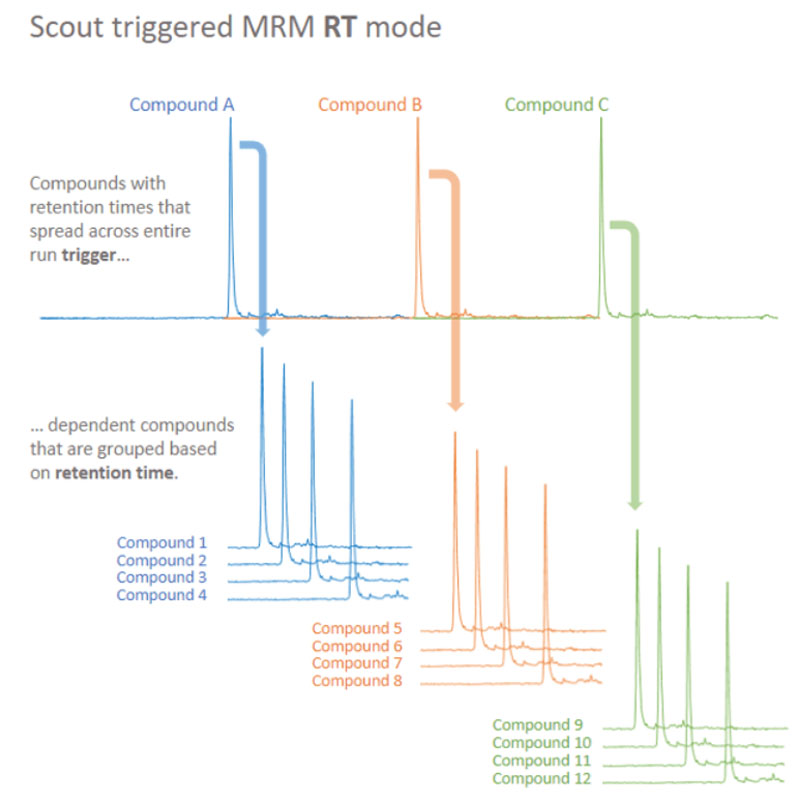
Figure 3: Principles of Scout-triggered MRM analysis. Scout-triggered MRM uses a marker-dependent transition to trigger MS analysis for a group of dependent target analytes. The marker-dependent transitions are typically staggered across the chromatographic run and dependent analytes are associated based on their retention times. The outcome is a targeted assay that is robust to retention time shifts8
When time-scheduling the acquisition of large numbers of MRM transitions, retention time windows have also become narrower to preserve data quality. This requires the MS operator to continuously monitor the results throughout the acquisition, for many samples and long periods of time.
Small changes to the chromatography system — such as mobile phase composition, gradient length and column phase — can cause retention times to shift. Consequently, the expected peaks of some analytes shift out of their scheduled retention time windows, resulting in analyte peaks not being acquired (Figure 2).8
Each target peak that is not acquired must be reacquired in another run. These reinjections and the manual adjustments required to align the retention times cost laboratories thousands of dollars in instrument time, precious sample volume and higher operating labour resources each year.8
There is also a need to develop quicker analytical methods that lead to faster clinical analyses; delays in identifying bacterial infections and thus optimal antibiotic therapy worsen patient outcomes and raise healthcare expenditures.3,4,10,11
Cutting-edge technology with Scout-triggered MRM
Professors Lemoine and Vandenesch worked with SCIEX on a new concept to improve MRM and developed an innovative and faster MRM method. Scout-triggered MRM (stMRM) removes the need to maintain accurate retention time windows for all analytes.8
Instead, stMRM relies on Scout marker compounds that serve to provide marker-dependent transitions that trigger the acquisition of dependent MRMs (Figure 3), meaning that each target analyte peak is captured despite any unanticipated retention time shifts.8
With stMRM, scientists have a new MRM mode that can be programmed with trigger thresholds for the marker-dependent transitions to define when an acquisition window begins (Figure 4).8 Exact expected retention times no longer need to be determined in advance. Elution order, however, is now a critical factor.8
The user interface of this operation mode has a built-in editing function for overlapping retention time windows. A marker-dependent transition that elutes close to the boundary of its acquisition window might risk missing its trigger if it elutes outside of that window.
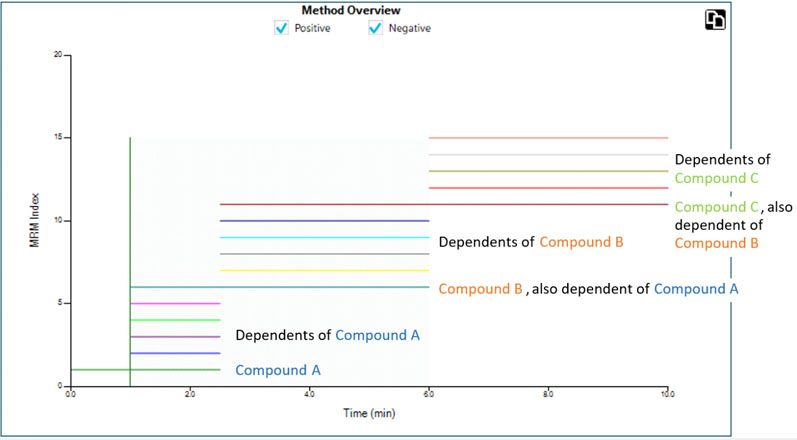
Figure 4: MRM transitions are triggered in acquisition windows across the chromatographic timescale. Users can programme the trigger thresholds for the marker dependent transitions to define when a window begins. Elution order is critical and exact retention times are no longer necessary8
To solve this issue, scientists can apply a retention time overlap function and apply a tolerance to each marker-dependent transition so that any adjacent transitions eluting within this tolerance will be triggered by that marker. By setting this tolerance, a dependent transition, for example, could be triggered by its own user-assigned marker or by the next or preceding one.8
Compared with the scheduled MRM algorithm, the Scout trigger MRM strategy allows for better optimisation of the acquisition windows to capture all target analytes — while retaining the same high standards for specificity, sensitivity, robustness and data quality (Figure 5).
Although this innovative new technology was only commercially launched this summer, Prof. Lemoine and SCIEX have already successfully assessed the analytical validity of stMRM by quantifying and evaluating candidate proteomic biomarkers for immune responses to pathogenic infection and environmental stress, and screening for multiresidue pesticides in surface water.12–14
Towards a future Scout-triggered MRM assay for clinical microbiology
Professors Lemoine and Vandenesch continue to work on providing the proof of concept to validate the LC-MS/MS MRM approach to rapidly identify and characterise pathogenic bacteria in clinical samples, including strains with AMR.3,4,15
For instance, they have successfully developed an MRM method that performs microbial identification, antibiotic-resistance detection and virulence assessment, as well as providing information for epidemiological typing within 60–80 minutes.3
They have also developed a prototype LC-MS/MS-based targeted proteomics assay using MRM to rapidly identify and characterise pathogenic bacteria from the endotracheal aspirate samples of patients with VAP.4
Assays such as these can provide a greater amount of actionable information from a biological sample in a single analytical run than other methods, such as PCR-based genotyping. Once the concept is proven, these assays could be refined for transition into a clinical setting with appropriate-grade equipment and reagents.
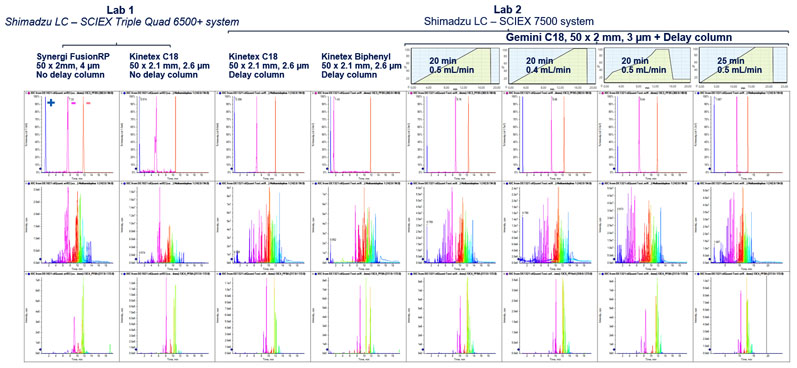
Figure 5: Testing the robustness of the Scout triggered MRM approach to varied chromatographic challenges. Despite changes to the LC system, such as adding a delay column, changing laboratories, and altering the flow rate, the Scout-triggered MRM workflow was able to trigger and detect the target analytes. The workflow is shown to overcome common method adjustments that affect retention time without requiring major rebuilding of the acquisition method8
Such data-rich tools could also shave off a significant amount of time from the bacterial identification and profiling workflow.3,4 Thus, physicians and patients could make better informed decisions earlier regarding which antimicrobial treatment to use to combat an infection.
With the World Health Organization (WHO) officially declaring AMR as one of the top 10 global public health threats facing humanity, it is imperative that we embrace multiple strategies to eliminate or radically minimise AMR by accurately and rapidly treating infections with effective medicine.16
Today, Prof. Lemoine’s latest research uses stMRM to monitor 300 characteristic peptides to specifically identify the pathogens involved in sepsis.15 This assay can analyse the 300 peptides in a mere 8 minutes.15 Although not yet ready for clinical application, this research could help to improve clinical diagnostic testing to support the precision prescription and regulated use of antibiotics for patients in as little as 3 years.15
References
- https://ec.europa.eu/info/law/better-regulation/have-your-say/initiatives/12963-Revision-of-the-EU-general-pharmaceuticals-legislation_en.
- www.whitehouse.gov/briefing-room/statements-releases/2021/06/13/carbis-bay-g7-summit-communique/.
- Y. Charretier, et al., “Rapid Bacterial Identification, Resistance, Virulence and Type Profiling using Selected Reaction Monitoring Mass Spectrometry,” Sci. Rep. 5, 13944 (2015): doi: 10.1038/srep13944.
- C. Bardet, et al., “Early and Specific Targeted Mass Spectrometry-Based Identification of Bacteria in Endotracheal Aspirates of Patients Suspected with Ventilator-Associated Pneumonia,” Eur. J. Clin. Microbiol. Infect. Dis. 40, 1291–1301 (2021).
- Antimicrobial Resistance Collaborators, “Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis,” Lancet 399, 629–655 (2022).
- A. Vasala, et al., “Modern Tools for Rapid Diagnostics of Antimicrobial Resistance,” Front. Cell Infect. Microbiol. 10, 308 (2020): doi: 10.3389/fcimb.2020.00308.
- S. Ayciriex, et al., “Streamlined Development of Targeted Mass Spectrometry-Based Method Combining Scout-MRM and a Web-Based Tool Indexed with Scout Peptides,” Proteomics 20, e1900254 (2020): doi: 10.1002/pmic.201900254.
- https://sciex.com/content/dam/SCIEX/tech-notes/technology/ruo-mkt-01-14731-a/Scout-triggered-MRM_Technology__RUO-MKT-02-14731-A.pdf.
- https://sciex.com/content/dam/SCIEX/pdf/tech-notes/all/mass-spectrometry-ScheduledMRM-0921010.pdf.
- K.K. Perez, et al., “Integrating Rapid Pathogen Identification and Antimicrobial Stewardship Significantly Decreases Hospital Costs,” Arch. Pathol. Lab. Med. 137, 1247–1254 (2012).
- D.K. Heyland, et al., “The Clinical Utility of Invasive Diagnostic Techniques in the Setting of Ventilator-Associated Pneumonia,” Chest 115, 1076–1084 (1999).
- B. Rougemont, et al., “Scout-MRM: Multiplexed Targeted Mass Spectrometry-Based Assay without Retention Time Scheduling Exemplified by Dickeya dadantii Proteomic Analysis during Plant Infection,” Anal. Chem. 89, 1421–1426 (2017).
- M. Leprêtre, et al., “From Shotgun to Targeted Proteomics: Rapid Scout-MRM Assay Development for Monitoring Potential Immunomarkers in Dreissena polymorpha,” Anal. Bioanal. Chem. 412, 7333–7347 (2020).
- A. Salvador, et al., “Scout-Multiple Reaction Monitoring: A Liquid Chromatography Tandem Mass Spectrometry Approach for Multiresidue Pesticide Analysis without Time Scheduling,” J. Chrom. A 1621, 461046 (2020): doi: 10.1016/j.chroma.2020.461046.
- https://sciex.com/about-us/press-releases/2022/sciex-launches-the-new-cornerstone-of-software-ecosystem-sciex-os-30.
- www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance.
