Metabolomics offers the opportunity to profile major genomic and epigenomic pathways, making it a crucial step in many areas of life science research.
From a clinical standpoint, metabolomics allows scientists to identify the metabolites responsible for critical pathways with greater confidence, as well as understand how they change when our physiological conditions alter, paving the way to highly targeted and personalised treatments for many metabolic and genetic disorders.
Novel separation techniques are enhancing the sensitivity, specificity and speed of metabolite measurement, thereby accelerating the pace of metabolomics research.
Ion mobility spectrometry (IMS) is a powerful analytical technique that has been widely applied in synthetic and analytical chemistry applications.
As interest in the separation, identification and quantification of biological samples (lipids, peptides, proteins and metabolites) developed, the potential of IMS coupled to MS (IMS-MS) was explored and, along with liquid and gas chromatography coupled to mass spectrometry (LC-MS and GC-MS), it was quickly adopted as part of the analytical “toolbox” for research in the field of metabolomics.
However, the comprehensive measurement of the metabolome is challenged by the limited analytical sensitivity and specificity of available instrumentation. This, combined with a lack of robustness in sample preparation methods, can lead to the selective loss of metabolites and ambiguity in metabolite annotation.

If metabolites can’t be well detected and measured with specificity in complex samples, it becomes difficult for researchers to understand what significance they hold with respect to physiological processes.1,2
When combined with the natural complexity of biological samples and the structural diversity of metabolites in living systems, these challenges directly affect the interpretation and downstream impact (translation) of analytical results.
Solving analytical challenges
The Metabolomics Platform group is one of five shared research facilities at the CBMR; its goal is to develop analytical strategies for metabolomics-driven systems biology.
The Moritz Group and other research teams at CBMR work together with colleagues in the Single-Cell Omics Platform Group, Computational Chemistry Unit, Phenomics Platform Group and Rodent Metabolic Phenotyping Platform Group to enable interdisciplinary research that transforms the basic understanding of the mechanisms involved in metabolic health and disease.
Using metabolomics, and often working in collaboration with other foundation-based scientists as well as external research groups, the Moritz Group focuses on answering questions related to the role of metabolism in disease, improved analytical strategies, better sample preparation methodologies and the appropriate use of analytical techniques.
The overall aim is to provide the knowledge that will lead to a greater likelihood of developing novel diagnostic procedures and therapeutics that will ultimately inform better clinical practice and improve the quality of life of affected patients.
The Moritz Group specialises in studying the metabolome in both large-scale analyses of human cohorts and in detailed investigations of metabolite flux in pathways (fluxomics). To achieve this, they devise novel methods to analyse and characterise metabolites.
Based on experience and a deep understanding of the available analytical solutions, they utilise the most appropriate combination of chromatography and MS technology to detect low concentrations of metabolites and characterise the composition of samples as accurately and comprehensively as possible.
This places an important responsibility on analytical instrument vendors to work in conjunction with researchers to deliver equipment and software solutions with the performance characteristics that enable progress to be made.
The development of TIMS — an analyte separation technique in which ions exhibit differential mobility according to their size and shape and complements mass analysers that separate ions according to their mass and charge state — is a key example of this in practice.3
TIMS yields a layer of separation and analytical specificity beyond that provided by mass analysis alone and by chromatography. For example, in a recent study, it was found that, when compared with a QTOF-only mode, TIMS visualised additional metabolite isomers and isobars, enabling imaging-based metabolomics with enhanced sensitivity and specificity at high spatial resolution.4
TIMS complements mass analysers that separate ions according to their mass and charge state and, in contrast to a conventional drift analyser in which the ions are passed through stationary gas, TIMS utilises a stream of moving gas and opposing electric fields to both trap and selectively release ions in the TIMS tunnel.3
The subsequent introduction of the Bruker vacuum-insulated probe heated electrospray ionisation (VIP-HESI) source in July 2021 gave improved sensitivity for a range of analyses.
For example, a key area of research for the Moritz Group concerns the analysis of the metabolism of small molecules; experiments using TIMS with a VIP-HESI source achieved dramatically higher resolution compared with conventional electrospray ionisation (ESI). Accurate mass measurement with exact isotropic patterns allowed the team to detect even minuscule amounts of metabolites in the samples (Figure 1).
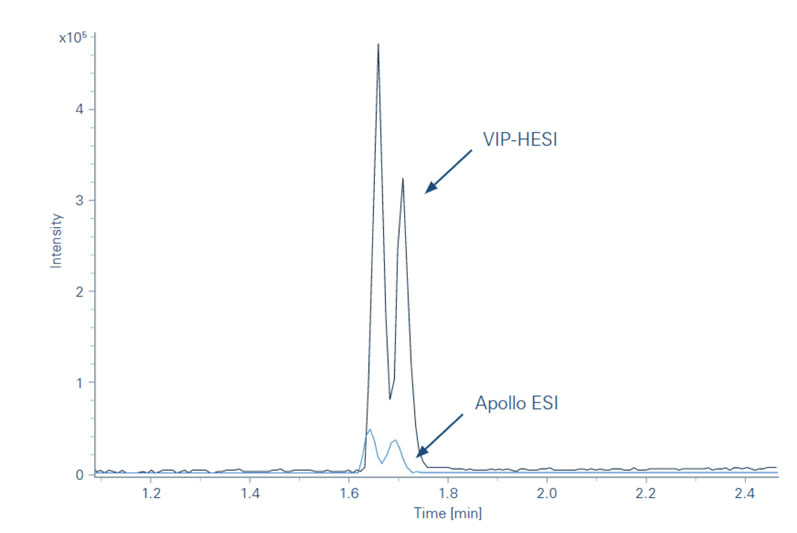
Figure 1: Illustration of the differences in metabolite sensitivity between VIP-HESI and conventional ESI
Novel research and new methods
Researchers at the CBMR have now started to use TIMS for different kinds of metabolic profiling. Two examples, taken from recently published work and presented below, show interesting results and raise the question of how TIMS would improve the metabolic profiling in such projects:
Exploring the NADome
Recent work focused on the measurement of nicotinamide adenine dinucleotide (NAD) and its metabolites (the so-called NADome). NAD is an essential coenzyme that is present in all organisms, serving as a major coenzyme for enzymatic reduction–oxidation reactions and adenosine triphosphate (ATP) production.
Most recently, NAD has been shown to be a crucial cosubstrate for numerous other enzymes. By identifying NAD levels within the blood, assessments can be made regarding whether or not the individual has a healthy metabolic function. If not, this could indicate the likelihood of a range of age-related or neurodegenerative diseases.5
In collaboration with other groups at CBRM, the Moritz Group has developed a more automated approach to sample preparation, along with new TIMS methodology that allows the analysis of a large cohort of thousands of samples. These represent essential steps in terms of decreasing labour-intensive preparation and reducing experiment times.
Understanding the link between molecular and phenotypic variability
Researchers from several European institutions came together in a recent study to evaluate how the human body responds to lifestyle coaching. Most data-driven health programmes fail to understand the association between molecular and phenotypic variability as the means to analyse these insights is limited.
Parameters were set such as clinical measurements, surveys, proteomics, genomics, metabolomics, autoantibodies and the gut microbiome. Researchers at the Moritz Group assisted with metabolomic experiments and interpretations of the data gathered.
Metabolites were identified using the retention index obtained from LC-MS and GC-MS systems. The dataset included 136 metabolites obtained from the GC-MS experiments, 104 LC-MS-negative and 163 LC-MS-positive metabolites verified against a predefined list of metabolites found in blood, plasma and serum.
Taking this into account, the researchers were able to integrate the data with datasets obtained for various other parameters (from different institutions). They concluded that there was a great degree of individual variation — between 56% and 71% on average — indicating high levels of variability between individuals.
The method also allowed profiling of the metabolites based on several parameters, such as dietary habits, hepatic function, physical exercise, sleep activity, BMI, etc., which are all influenced by the presence of specific metabolites.
Ultimately, the method allowed them to prove that metabolic pathways differ between individuals based on genetic factors, habits, and current physical state.6
One wonders, however, how much more valuable data would have been acquired by using a TIMS-QTOF approach rather than a QTOF method, which offers more opportunities to separate and distinguish between isomeric and isobaric compounds and higher annotation certainty.
Looking ahead
With the importance of metabolomics firmly established, and the significant insight it can give for molecules that are responsible for critical biological pathways and are implicated in major diseases proven, research efforts are turning to
- improving analytical strategies for efficient measurement
- integrating metabolomics data with other ‘omics information, healthcare records and lifestyle data.
The perspectives and research introduced here by the Moritz Group suggests that significant progress is being made — both for understanding basic science and for the advancement of precision medicine.
References
- A. Di Minno, et al., “Challenges in Metabolomics-Based Tests, Biomarkers Revealed by Metabolomic Analysis and the Promise of the Application of Metabolomics in Precision Medicine,” International Journal of Molecular Sciences 23(9), 5213 (2022): doi: 10.3390/ijms23095213.
- D.S. Wishart, “Emerging Applications of Metabolomics in Drug Discovery and Precision Medicine,” Nature Reviews Drug Discovery 15, 473–484 (2016).
- M.E. Ridgeway, et al., “Trapped Ion Mobility Spectrometry: A Short Review,” International Journal of Mass Spectrometry 425, 22–35 (2018).
- E.K. Neumann, et al., “Spatial Metabolomics of the Human Kidney using MALDI Trapped Ion Mobility Imaging Mass Spectrometry,” Analytical Chemistry 92(19), 13084–13091 (2020).
- S. Lautrup, et al., “NAD+ in Brain Aging and Neurodegenerative Disorders,” Cell Metab. 30(4), 630–655 (2019).
- F. Marabita, et al., “Multiomics and Digital Monitoring During Lifestyle Changes Reveal Independent Dimensions of Human Biology and Health,” Cell Systems 13(3), 241–255 (2022).
