Some of the current bottlenecks with mRNA-based drugs is their short half-life because of rapid degradation and high dosage requirements. This is likely to result in the need for subsequent administrations that approach or exceed acceptable ranges of impurities for certain indications.
Thanks to their built-in copy mechanism, which expresses the proteins responsible for replication derived from viral genomes, self-replicating RNAs (srRNAs) can address this challenge. Thus, a higher quantity of the target protein can be produced from a single RNA dose.
The emergence of srRNA drugs has positive implications for nucleic acid-based therapies and vaccines, such as reduced efficacious human dosage, the potential for increased efficacy and safety, faster manufacturing and lower production costs.
It also enables additional modes of action, such as T-cell activation, which could lead to more effective vaccines and anticancer therapeutics. However, the built-in copy machinery of srRNAs requires a significant increase in size, making the analysis of srRNA more complicated.
Capillary electrophoresis (CE), which has previously been used for various RNA separation applications, emerges as a promising tool to evaluate the integrity and purity of these large RNAs.
This article covers the benefits and challenges of srRNAs and how CE technology can contribute to the srRNA development.
In-vitro transcribed drugs
Nucleic acid-based drugs are becoming increasingly prominent for the treatment or prevention of several conditions.

Initial nucleic acid therapeutics were based on synthetically produced oligonucleotides, such as antisense oligonucleotides, small interfering RNAs (siRNAs) and microRNAs (miRNAs), to name a few.
In contrast, IVT products rely on a plasmid that gets transcribed in vitro into RNA. Upon administration, IVT RNAs can transiently induce the expression of a protein of interest without entering the host nucleus.
Thus, unlike plasmid-based drugs and viral vectors, IVT RNAs do not pose the risk of integrating into the patient’s genome and causing mutations.
The functionality of IVT RNAs has been recently demonstrated in SARS-CoV-2 vaccinations, whereby mRNA was delivered to patients’ cytoplasm to encode target antigens that stimulate antibody production.
Current mRNA-based SARS-CoV-2 vaccines on the market or in clinical trials comprise non-replicating mRNA.1 mRNA in vaccines exhibits its effects transiently and is degraded shortly after inducing the desired immune response.
To optimise the stability and translation efficiency of IVT mRNA and avoid undesired immune reaction, nucleoside modifications were introduced into mRNA.
Two of the current mRNA vaccines for COVID-19, Spikevax from Moderna and Comirnaty from Pfizer/BioNTech, contain a modification of uridine into N1-methyl pseudouridine (1MpU) to enhance antigen expression and improve stability.2
Self-replicating RNA to extend protein production: positive implications
Although a sharp and short-lived peak in target protein production could be achieved with mRNAs and base-modified mRNAs (bmRNAs), the challenge remains to prolong substantial protein production with minimal dosage.
mRNA half-life is one of the main obstacles for the successful application of mRNA drugs to cancer and autoimmune diseases. Conventional mRNA technology would require the repeated administration of high doses to achieve therapeutic effects, which poses the risk of adverse events.
Additionally, this puts pressure on manufacturers in terms of input material costs and large production outputs.
srRNA has emerged as a promising approach because of its ability to make copies of itself following cellular uptake.
In particular, it was discovered that positive-strand RNA viruses possess the ability to complete a replication cycle owing to the built-in machinery responsible for the production of proteins needed for replication.3
Researchers leveraged this mechanism from RNA viruses and derived srRNA that was able to not only code the target protein, but also the proteins involved in replication.
Specifically, srRNAs entering a host cell express the proteins that facilitate RNA replication, such as RNA-dependent RNA polymerase. The RNA copies can then be translated into the target protein by the ribosome of the host cell.
Ultimately, srRNA administration drastically increases the protein output as well as durability.
srRNA offers a double-layered advantage compared with mRNA by improving treatment outcomes and reducing the minimum effective dose. In the case of vaccine production, the main upgrade is the reduced RNA dosage requirement that allows delivery to a much larger population in unit time.
If a vaccine can stimulate the desired immune response with a lower RNA dose, the same volume of batch production can be manufactured quicker, using less resources and distributed to many more individuals.
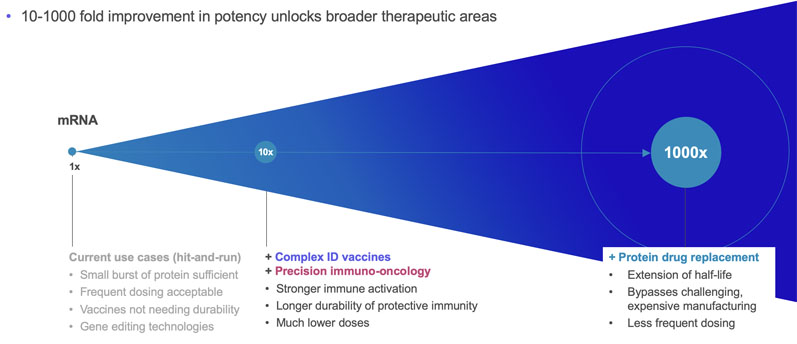
srRNA vector optimisation drives mRNA outperformance
The resulting cost reduction and faster turnaround can provide significant value by enabling prompt responses to future pandemics.
The lower dosing also has positive implications for the treatment of diseases with RNA-based therapeutics. Currently, administering the minimum effective mRNA dose would expose the patient to severe dose-dependent adverse effects for certain indications.
Those are caused by patients’ innate immune responses to the foreign RNA encapsulated in a lipid nanoparticle and impurities that can be present. Therefore, decreasing the initial RNA dosage with srRNAs can reduce adverse effects.
The innovative value of srRNA technologies can impact the outcomes of treatments for cancer, infectious diseases and autoimmune diseases, whereby a single dose can exhibit a prolonged therapeutic effect and adhere to safety regulations.
srRNAs are becoming increasingly important in terms of saving patient lives and/or improving prevention through vaccines.
As of 2022, various srRNA drug modalities have entered clinical trials for oncology, rabies and SARS-CoV-2, while preclinical studies of many other srRNA drugs are under way for cancer, HIV and infectious diseases (such as influenza, Ebola, malaria, and respiratory syncytial virus).4,5
Replicate Bioscience is one of the companies at the forefront of srRNA research; their goal, by amplifying target protein production, is to reduce the required doses by more than a thousand-fold and unlock new opportunities to treat patients across a broad range of diseases, including cancer.
The company is a pioneer in developing srRNA drugs and is conducting preclinical studies for cancer subtypes that are resistant to conventional therapies. When addressing the unmet needs in such complex diseases, the overall quality of the final srRNA product is of utmost importance.
Dr Andrew Geall, cofounder and chief development officer at Replicate Bioscience, expects that advancements in research and related analytics can unlock srRNAs up to 16,000 bases and encode multiple genes.
Although RNAs of that length have historically been challenging to manufacture, a particular separation method — capillary electrophoresis — may be suitable for the assessment of integrity and purity.
“Our first programme with a 16 kb construct is in oncology. Although still in the earlier stages, our analytical strategy of using CE combined with in vitro potency assays has been performing as expected and informing productive initial discussions with the US FDA."
"Right now, we want to show that it's possible to manufacture and analyse a 16 kb construct. This was inconceivable a few years ago,” says Andrew.
The challenges in analysing large srRNAs
Although srRNA technology could tackle the main challenges and indications of current RNA-based drugs, analytical assays in this field are in their infancy.
One hurdle that is evident with srRNA analysis is the significant increase in size and molecular weight of these therapeutics — owing to the additional encoding for proteins required for self-amplification.
Generally speaking, srRNAs can comprise up to 16,000 bases, whereas mRNA usually has a length of 2000–4000 bases.6
In contrast, current strategies are mostly suited to the analysis of these smaller mRNA modalities. This includes the availability of standards and ladders used to enable accurate assessment of these molecules.
Collaboration between vendors and pioneering biotech companies is critical. Andrew comments: “There is currently no standard beyond 9000 bases. We're working with vendors and providers to generate a reference material that the scientific community can access.”
Furthermore, satisfying regulatory requirements for the approval of srRNA as a drug cannot necessarily be achieved in the same way as for other nucleic acid-based drugs.
Analytical approaches need to be revised and adjusted, which can lead to a prolonged process to prove compliance and extend the time-to-market.
Robust analytical solutions are key to assessing quality parameters such as integrity and purity. This holds especially true for srRNAs with their larger size, which complicates the analysis of these critical parameters.
Capillary electrophoresis and its value in srRNA development
Separation techniques are integral to establishing the robustness and control of the production of high-quality RNAs and other therapeutic drugs. CE can be invaluable owing to the plethora of separation methods it offers as well as being the preferred mode of analysis for mRNA purity.7
CE assays employed in the mode of capillary gel electrophoresis (CGE) can fulfil the criteria for srRNA analysis, such as high separation efficiency and baseline resolution between analyte peaks with minimal sample volume requirements.
Degradation by-products, which cause size and purity heterogeneity, can be robustly detected using CGE. For example, a CGE analysis in denaturing conditions can disrupt secondary structures so that separation is based primarily on the number of nucleotides.7
Thus, CGE-mediated IVT RNA sample analysis can yield sharp, well-defined peaks that facilitate high-performance separation.
CE manufacturers must continuously upgrade the capabilities of their solutions to fulfil the latest biopharma research and development demands, such as a vast increase in molecule size (as is the case for srRNA).
The launch of the RNA 9000 Purity & Integrity kit by SCIEX set a milestone for single-stranded nucleotide analysis, covering the largest size range from 50–9000 nucleotides.
To support the biopharma community, SCIEX listens to customer problems and continuously works to expand its solution portfolio to achieve the maximum resolution in the fastest time frames with kit-based workflows.
By implementing advanced and innovative CE solutions, more detailed characterisation can be enabled and regulatory compliance can be achieved much more efficiently.
By undertaking product evolution in CE solutions, instrument manufacturers will be indisputably valuable to biopharma and biotech working on srRNAs allowing to respond quickly and efficiently to global health crises.
References
- E. Fang, et al., “Advances in COVID-19 mRNA Vaccine Development,” Sig. Transduct. Target. Ther. 7, 94 (2022).
- D.M. Mauger, et al., “mRNA Structure Regulates Protein Expression Through Changes in Functional Half-Life,” Proceedings of the National Academy of Sciences 116(48), 24075–24083 (2019).
- M-C. Bernard, et al., “The Impact of Nucleoside Base Modification in mRNA Vaccine is Influenced by the Chemistry of Its Lipid Nanoparticle Delivery System," Molecular Therapy-Nucleic Acids 32, 794–806 (2023).
- T. Lu, et al., “High-Resolution Capillary Electrophoresis Separation of Large RNA Under Non-Aqueous Conditions,” Journal of Chromatography A 1618 (2020): 460875Tews
- A. Birke and G. Meyers, “Self-Replicating RNA,” Methods in Molecular Biology 1499, 15–35 (2017).
- L. Schoenmaker, et al., “mRNA-Lipid Nanoparticle COVID-19 Vaccines: Structure and Stability,” International Journal of Pharmaceutics 601, 120586 (2021).
- Z. Li, et al., “Capillary Electrophoresis of RNA in Hydroxyethylcellulose Polymer with Various Molecular Weights,” Journal of Chromatography B 1011, 114–120 (2016).
